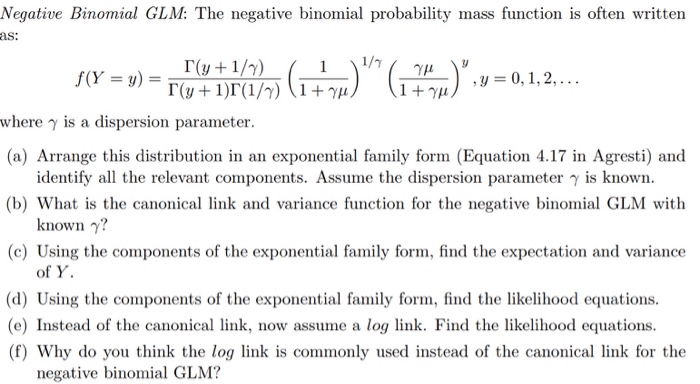
Glm Negative Binomial Link Function - With the logit or log-odds link function gx lnleftfracx1-xright the likelihood for a single observation becomes. Selecting Link Function for Negative Binomial GLM.
Http Friendly Apps01 Yorku Ca Psy6136 Lectures Countdata 4up Pdf
Note that while phi is the same for every observation y_i and therefore does not influence the estimation of beta the weights w_i might be different for every y_i such that the estimation of beta depends on them.

Glm negative binomial link function. Logit probit and complementary log-log. Therefore it is said that a GLM is determined by link function g and variance function vmu alone and x of course. Judging from a quick look it seems to me like he only uses the log link.
Glmrespvar familybinomiallinklogit Or does it use a different link as default. The negative binomial requires the use of the glmnb function in the MASS package. A modification of the system function glm to include estimation of the additional parameter theta for a Negative Binomial generalized linear model.
The call to glmnb is similar to that of glm except no family is given. The available link functions are Link function glm option identity linkidentity log linklog logit linklogit probit linkprobit complementary loglog linkcloglog odds power linkopower power linkpower negative binomial linknbinomial loglog linkloglog log-complement linklogc. To fit a negative binomial model in R we turn to the glmnb function in the MASS package a package that comes installed with R.
Again we only show part of the summary output. The negative binomial distribution belongs to the exponential family and the canonical link function is. The link function converts the LS-.
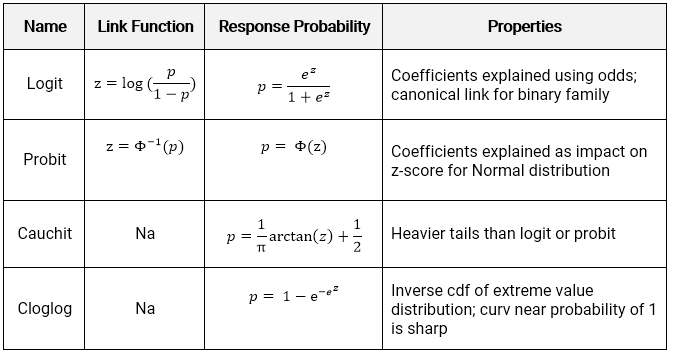
The link function converts the model scale LS-mean estimates back to the original data scale. The two most common link functions used for binomial GLMs are the logit and probit functions. Usage glmnbformula data weights subset naaction start NULL etastart mustart control glmcontrol method glmfit model TRUE x FALSE y TRUE contrasts NULL inittheta link log.
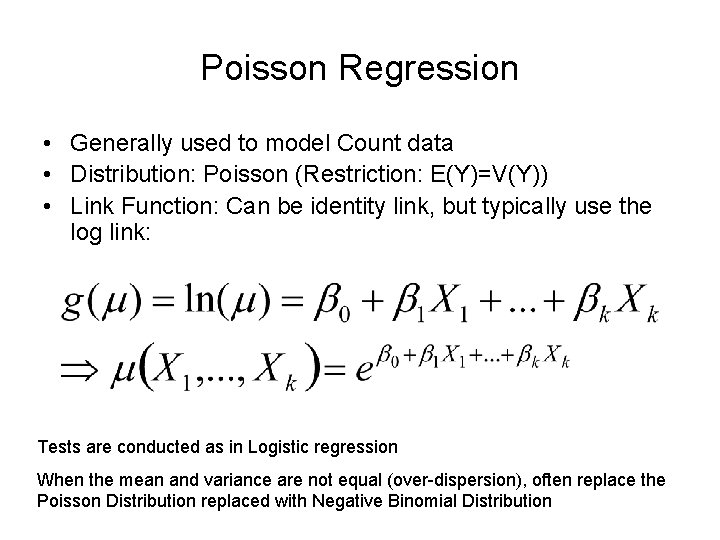
Proc glimmix uses a distribution to estimate model parameters. In Poisson and negative binomial glms we use a log link. G μ i log.
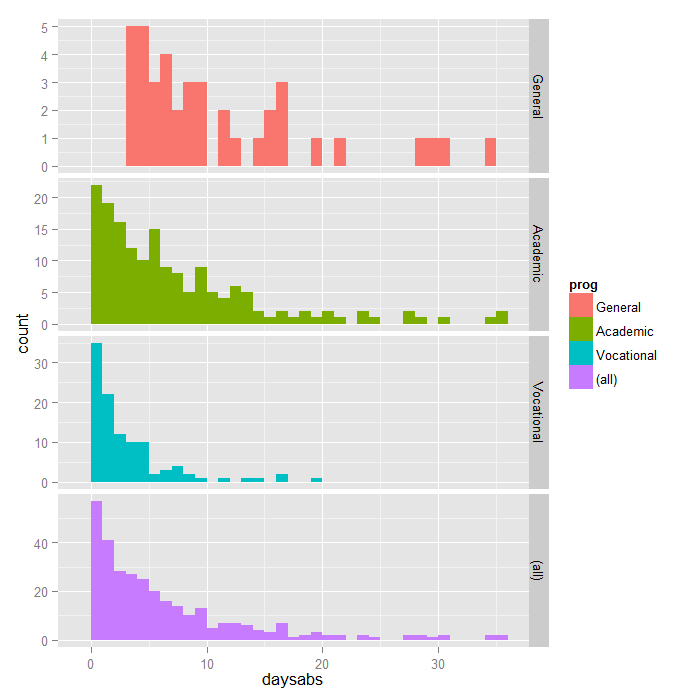
The data values are not transformed by the link function. We noticed the variability of the counts were larger for both races. The probability function defines the Negative Binomial distribution.
μ i k μ i log. Yi i Poisson i D-20. Gμi ηi α β1Xi1 β2Xi2 βkXik 1Some authors use the acronym GLM to refer to the general linear modelthat is the linear regression model with.
Logit is the default choice. It is a discrete distri-bution frequently used for modelling processes with a response count for which the data are overdispersed relative to the Poisson distribution. Two key concepts are.
I dont recall having seen any other links in practice. Im trying to model insect abundance data with a variety of vegetationsite related covariates. The log is just so very easy to work with - it gives positive parameter values its differentiable and easily so.
It does not fit the data to a distribution. The actual model we fit with one covariate x looks like this Y sim textPoisson lambda loglambda beta_0 beta_1 x here lambda is the mean of Y. Specifies the information required to fit a Negative Binomial generalized linear model with known theta parameter using glm.
There are three common choices for link functions regarding binomial data. Enter the following commands in your script and run them. The standard reference IMO is Hilbe Negative Binomial Regression.
The log link is usually preferred because of the analogy with Poisson model and it also tends to give better results. So if we have an initial value of the covariate x_0 then the predicted value of the mean lambda_0 is given by. A smooth and invertible linearizing link function g which transforms the expectation of the response variable μi EYi to the linear predictor.
K μ i 1 but it is difficult to interpret. Family by using family. Usage negativebinomialtheta stoptheta must be specified link log Arguments.
Poisson and negative binomial GLMs. Using the notation described in Equation D-15 the NB2 model with spatial interaction can be defined as. Because it is count data that is over-dispersed Ive decided to use the negative binomial distribution.
It would appear that the negative binomial distribution would better approximate the distribution of the counts. To capture this kind of data a spatial autocorrelation term needs to be added to the model. Family function for Negative Binomial GLMs Description.
The Poisson-Gamma or negative binomial model can also incorporate data that are collected spatially. Membership of the GLM family The Negative Binomial distribution belongs to the GLM family but only if the parameter.
Some Link Functions Used In Glm And Glmm And Their Interpretations Download Table
Negative Binomial Glm The Negative Binomial Chegg Com
Negative Binomial Regression R Data Analysis Examples
Some Distributions Used In Glm And Glmm Link Functions And Download Table
Negative Binomial Models Ppt Video Online Download
Http Friendly Apps01 Yorku Ca Psy6136 Lectures Countdata 4up Pdf
Count Models Understanding The Log Link Function The Analysis Factor
Possible Combinations Of Link Functions And Distributions In Xthybrid Download Table
Generalized Linear Models Generalized Linear Models Glm General
Linear Structure And The Link Function Sage Research Methods
10 Glms For Classification Exam Pa Study Guide Fall 2021
Use With Generalized Linear Models Sage Research Methods
Generalized Linear Models Ii Distributions Link Functions Diagnostics
Exotic Link Functions For Glms Freakonometrics