Glm Negative Binomial Stata - Generalized linear models GLMs provide a powerful tool for analyzing count data. - You will need to use the glm command to obtain the residuals to check other assumptions of the negative binomial model see Cameron and Trivedi 1998 and Dupont 2002 for more information.
How To Produce A Graph Of Predicted Values Versus Observed Values After An Overdispersed Poisson Regression
15 that SPSS included GLM procedures while Stata offered included negative binomial regression in its GLM package in the early 90s over a decade earlier.

Glm negative binomial stata. In glm you can specify familynbinomial _k and then search for a _k that makes the deviance-based dispersion equal to 1. Its default value is 1 so if alpha is not 1 the results will be different. The negative binomial distribution belongs to the exponential family and the canonical link function is.
I believe the code for negative binomial regression is glm x y familynbinomial linklog but I do not know how to indicate truncation. Usage glmnbformula data weights subset naaction start NULL etastart mustart control glmcontrol method glmfit model TRUE x FALSE y TRUE contrasts NULL inittheta link log. K μ i 1 but it is difficult to interpret.
Once again we can exponentiate the race coefficient to get a ratio of sample means and make predictions to get the original sample means. However you can also use familynbinomial ml to estimate _k with maximum likelihood which should report the same value as nbreg. It wasnt until v.
Fmeans fmeans same as pGLM 1. The traditional negative binomial regression model commonly known as NB2 is based on the Poisson-gamma mixture distribution. The reason this works is that the negative binomial distribution with a known theta parameter falls in the exponential family which means that standard GLM-fitting techniques such as IRLS will work fine.
Thank you for your assistance. The following code shows how to fit both a Poisson regression model and negative binomial regression model to the data. Nb offers division exam data data.
Fitted counts for Negative Binomial GLM. The log link is usually preferred because of the analogy with Poisson model and it also tends to give better results. 4glm Generalized linear models By default scale1 is assumed for the discrete distributions binomial Poisson and negative binomial and scalex2 is assumed for the continuous distributions Gaussian gamma and inverse Gaussian.
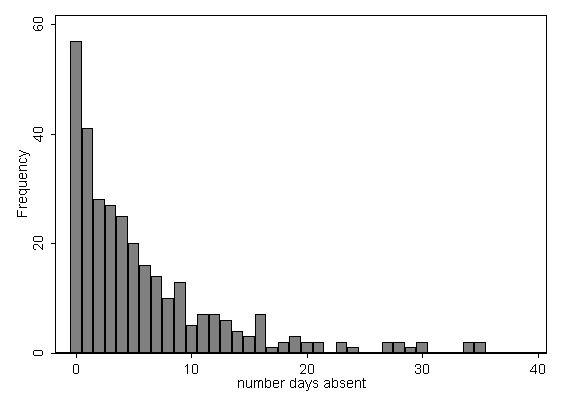
Negative binomial regression is a generalization of Poisson regression which loosens the restrictive assumption that the variance is equal to the mean made by the Poisson model. Sum_i1n_2 Y_i - hatY_i2. Negative binomial models in R are limited as of this writing but more advanced models are sure to follow in the near future.
1 The starting point for count data is a GLM with Poisson-distributed errors but not all count data meet the assumptions of the Poisson distribution. On the other hand nbreg will also give you a confidence interval. - You can also run a negative binomial model using the glm command with the log link and the binomial family.
A modification of the system function glm to include estimation of the additional parameter theta for a Negative Binomial generalized linear model. The K parameter requested by GLM is exactely the alpha parameter of negative bin regression and it is equal to 1k. Glm fits generalized linear models.
You can definitely use glm to fit this model. Fit Poisson regression model p_model. See U 27 Overview of Stata estimation commands for a description of all of Statas estimation commands several of which fit models that can also be fit using glm.
Now build both the Poisson model and the negative binomial model based on your training data set. Calculate the predicted values for the data in your testing data set and compare it to the actual values in the following way. Scalex2 specifies that the scale parameter be set to the Pearson chi-squared or generalized chi-.
G μ i log. Choosing among Poisson negative binomial and zero-inflated models Ecologists commonly collect data representing counts of organisms. All data sets and Stata ado files related to models used in the text can be.
Is it possible to use the GLM command for zero truncated negative binomial regression. μ i k μ i log. It can fit models by using either IRLS maximum quasilikelihood or NewtonRaphson maximum likelihood optimization which is the default.
Count data and GLMs. The inverse of the first equation gives the natural parameter as a function of the expected value θ μ such that. With v μ b θ μ.
Therefore it is said that a GLM is determined by link function g and variance function v μ alone and x of course. 2 Doug is correct that if you have GLM machinery working the way to estimate the negative binomial model is to optimize over the theta parameter.
Different Results For Negative Binomial Regression For Dispersion Mean Constant Statalist
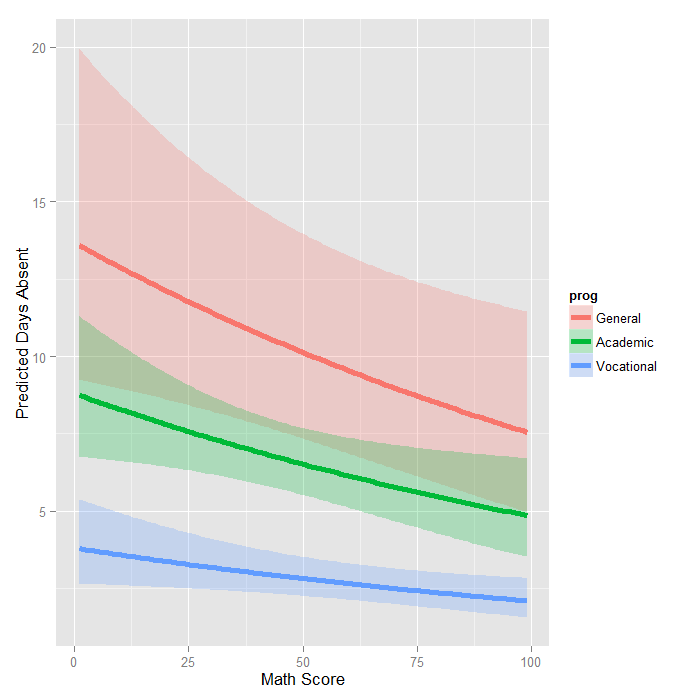
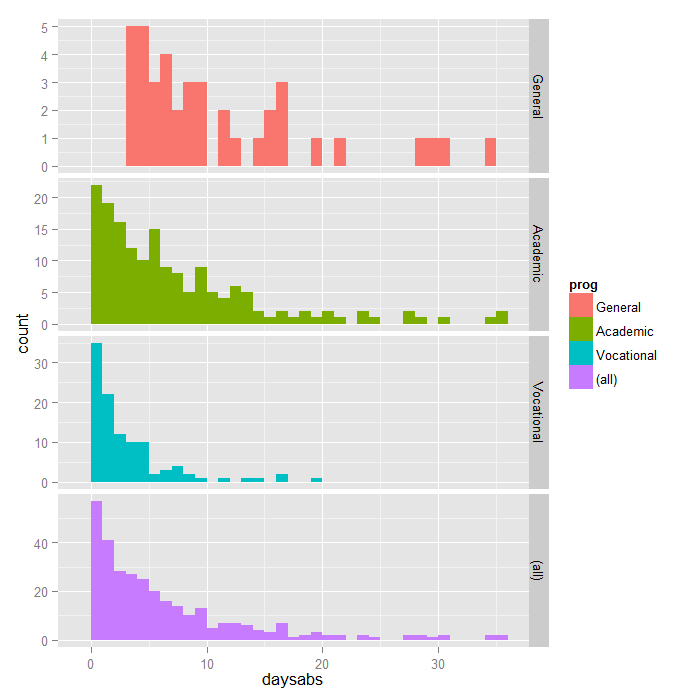
Negative Binomial Regression R Data Analysis Examples
How To Choose Between Poisson Regression Model And Negative Binomial Model Statalist
Glm Model Fit Can T Find A Family Link Combination That Produces Good Fit Cross Validated
Negative Binomial Regression Stata Data Analysis Examples
Poisson Model Versus Negative Binomial Model With Equidispersion Statalist
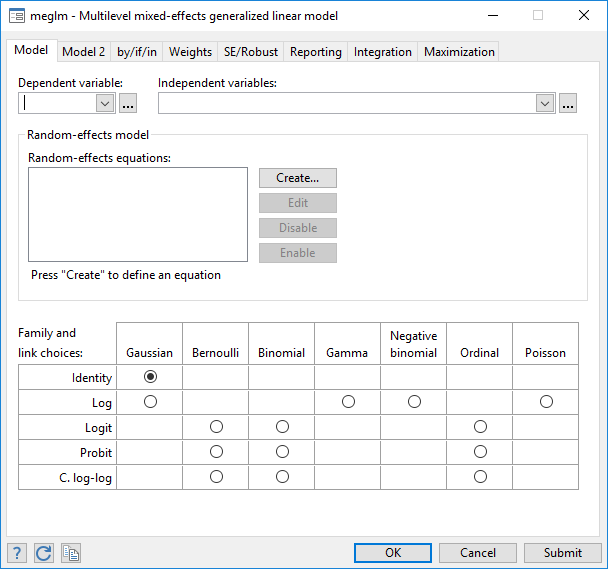
Multilevel Generalized Linear Models Stata
Getting Started With Negative Binomial Regression Modeling University Of Virginia Library Research Data Services Sciences
Different Results For Negative Binomial Regression For Dispersion Mean Constant Statalist
Stata Bookstore Negative Binomial Regression Second Edition
Negative Binomial Regression R Data Analysis Examples
Glm In R Negative Binomial Regression V Poisson Regression Youtube
Negative Binomial Regression Stata Data Analysis Examples
How To Choose Between Poisson Regression Model And Negative Binomial Model Statalist